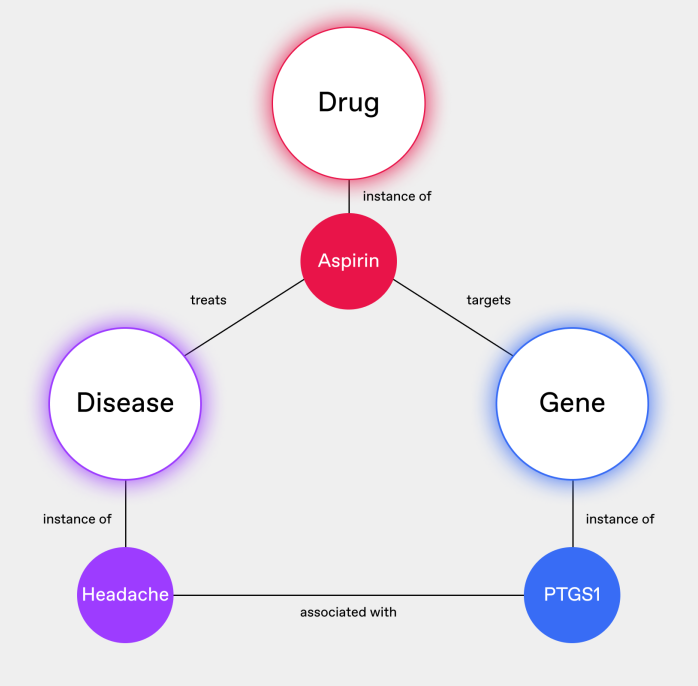
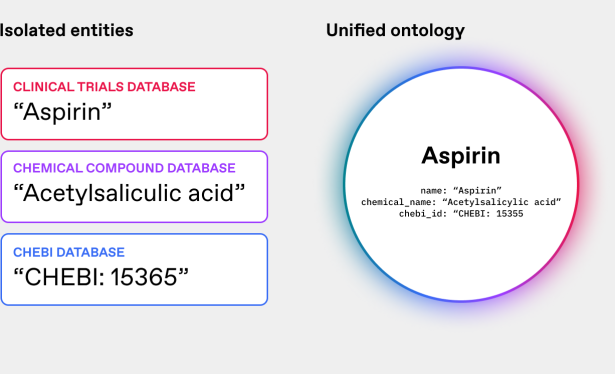
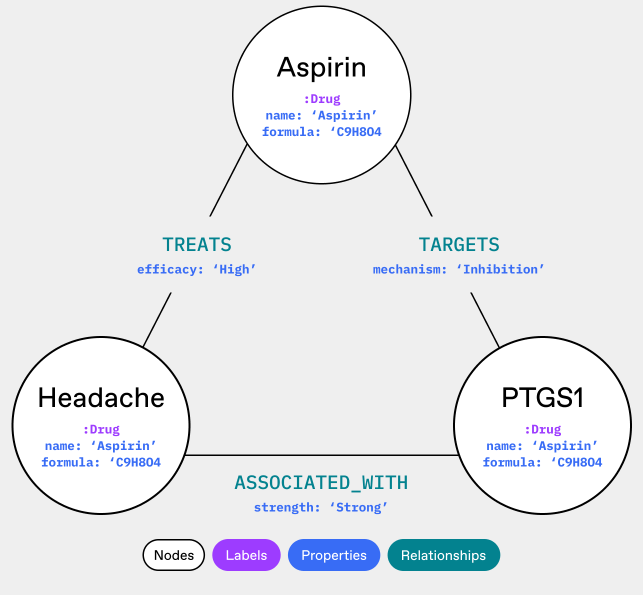
An ontology can define that all these terms refer to the same entity (Fig. 2), allowing researchers to pull comprehensive information about the drug from multiple sources without missing crucial data or inadvertently treating them as separate compounds.
This connective power extends beyond just naming conventions. Ontologies can also bridge gaps in the granularity of concepts across datasets. One database might refer broadly to "NSAIDs" (Non-Steroidal Anti-Inflammatory Drugs), while another lists specific drugs like Aspirin, Ibuprofen, and Naproxen. A well-designed ontology can establish the hierarchical relationship between these concepts, enabling researchers to navigate between general classes of drugs and their specific instances.
As you can see, ontologies enable researchers to integrate data from diverse sources confidently, perform more comprehensive analyses by ensuring no relevant data is overlooked, and discover non-obvious relationships between entities that might be obscured by inconsistent naming or classification.
Building your ontology
When embarking on the journey to create an ontology, one of the first crossroads you'll encounter is whether to use an existing public ontology or build your own. This decision is critical and can significantly impact the effectiveness and efficiency of your knowledge graph.
Evaluating public vs. custom ontologies
Public ontologies like ChEBI, RxNorm, and ClinicalTrials.gov have the advantage of being well-established and widely recognized in the scientific community. They've been refined over years and offer a common language that facilitates data sharing and collaboration. However, they may only sometimes perfectly align with your research needs or data structures.
On the other hand, building your ontology allows you to tailor it precisely to your requirements. You can define classes and relationships directly corresponding to your data and research focus. But this path comes with challenges - it requires significant time, expertise, and resources.
The key to making this decision is understanding how well existing ontologies map to the datasets you care about. This involves a deep evaluation of your data sources, research objectives, and the capabilities of public ontologies. You might find that a combination of public and custom ontologies best serves you, allowing you to leverage established standards while accommodating your unique needs.
Understanding existing ontologies
Before you decide to build your own ontology from scratch, it's crucial to understand what's already out there.
Which public ontologies in your domain are the big players?
How well do these ontologies cover the concepts and relationships in your data sets?
Are existing ontologies too zoomed-out or too zoomed-in for what you need?
How often are public ontologies updated? You want to rely on something other than outdated information.
This step is gold - it could save you a ton of time and effort if an existing ontology (or a clever combo of a few) fits the bill.
Going with your own ontology
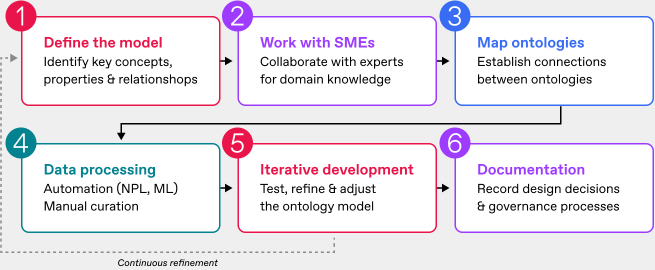
Creating your own ontology demands some important steps.